
|
RNA-Seq |
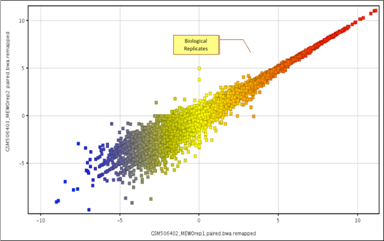
|
Quantification and Normalization
Expression values at gene, exon and transcript level. RPKM, DESeq, Quantile and TMM methods for normalization. |
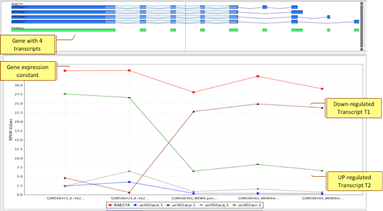
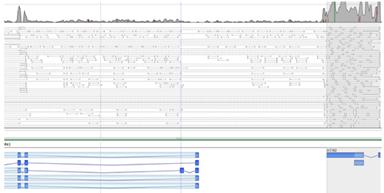
Differential SplicingDetermine gene and deconvoluted transcript expression profiles using EM algorithm; identify alternative splicing patterns. |
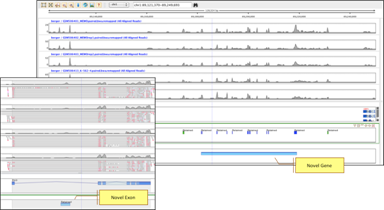
Novel DiscoveryAnalyze coverage patterns to detect novel genes and exons not present in NCBI, UCSC, and Ensembl annotations. |
|
Differential Expression
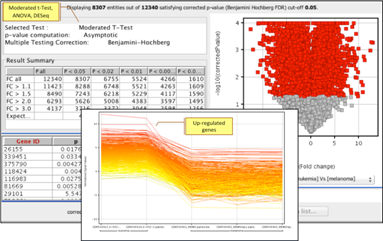
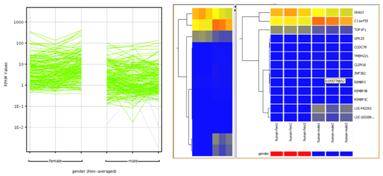
t-Test, Mann-Whitney, n-way ANOVA, and DESeq for identifying differentially expressed genes under different experimental conditions. Multiple testing correction using Bonferroni and Benjamini Hochberg methods. |
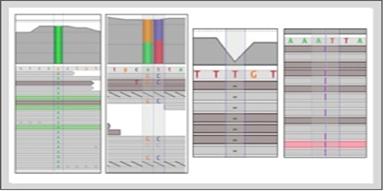
Transcriptome VariantsIdentify variants in transcriptome, predict effect on transcripts, and perform differential SNP analysis. |
Gene FusionFind gene usions via spliced and paired reads spanning the fused genes. Results annotated with paralogs and pseudogenes information for identifying false positives. |
Clustering GenesCluster genes on their expression values to identify groups of genes behaving similarly across conditions. |
|
Strand NGS supports an extensive workflow for the analysis and visualization of RNA-Seq data. The workflow includes standard differential expression analysis for different experimental conditions, as well as differential splicing analysis. It supports novel discovery including identifying novel genes and exons and novel splice junctions. It includes the ability to detect variants in the transcriptome, and the ability to detect gene fusion events. Further downstream analysis such as GO, pathway analysis, etc can be performed on the set of interesting genes. |

