
|
small RNA-Seq |
Comprehensive AnnotationsComprehensive Small RNA annotations and structures including miRNA, tRNA, snRNA, and snoRNA from multiple sources. |
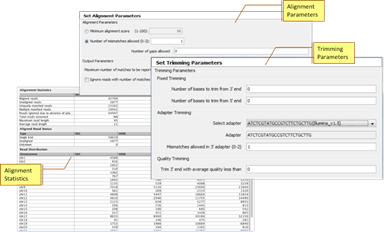
AlignmentSupport for alignment using the in-built Strand NGS alignment algorithm, with support for adapter trimming including adapter mismatches. |
|
Strand NGS supports an extensive workflow for the analysis of small RNA data. It provides the ability to determine the expression levels of various small RNA species. It supports detection of novel small RNA genes, and prediction of their type. It provides the ability to identify differentially expressed small RNA genes, and visualize the results in a small RNA-specific gene view. The mRNA targets of interesting small RNA genes can be identified from multiple target prediction databases. |
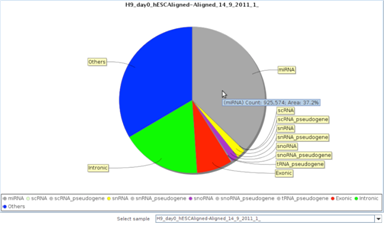
Quality ControlAnnotation based quality control visualizations in addition to read level quality plots and filters. |
QuantificationFind the expression levels of small RNA genes and mature miRNAs. Normalize the read counts using DESeq, TMM, or Quantile normalization. Visualize results in the small RNA Gene View. |
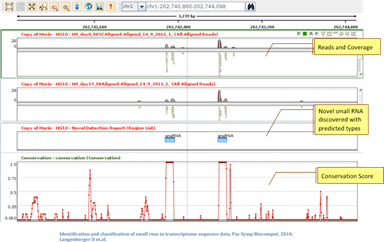
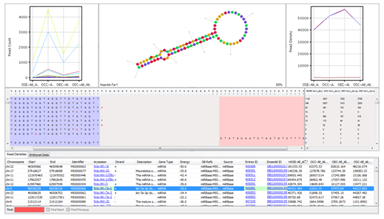
Novel Small RNA discoveryDetect novel small RNA genes and classify them as miRNA, snoRNA, scRNA, or tRNA. Identify high-confidence predictions with conservation scores and confidence values. Find annotation discrepancies of known genes from the read coverage patterns. |
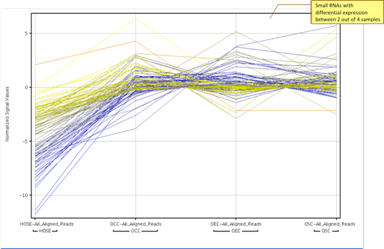
Differential Expressiont-Test, Mann-Whitney, n-way ANOVA, and DESeq for identifying differentially expressed genes under different experimental conditions. Multiple testing correction using Bonferroni and Benjamini Hochberg methods. |
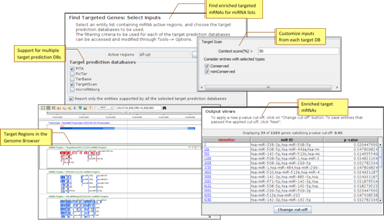
miRNA Target AnalysisIdentify protein-coding genes targeted by mature miRNAs based on mappings from various target prediction databases-TargetScan, PicTar, TarBase, microRNA.org, PITA. |

