

|
ChIP-Seq |
Peak DetectionIdentify protein binding sites using either Enriched Region Detection, PICS, or, MACS peak finding algorithms. View the results in the Genome Browser the context of known genes. |
|
Strand NGS supports a workflow for the analysis and visualization of ChIP-Seq data. This workflow provides the ability to identify transcription factor binding sites and histone modification sites using either Enriched Region Detection, PICS, or MACS peak detection algorithms. It supports the ability to detect motifs in the peak regions using GADEM, and scan for known motifs in the genome or regions of interest. Normalized read coverage for the Transcription Start Sites can be viusalized using the TSS Plot. Further, downstream analysis such as GO, pathway analysis, etc. can be performed on the set of affected genes. |
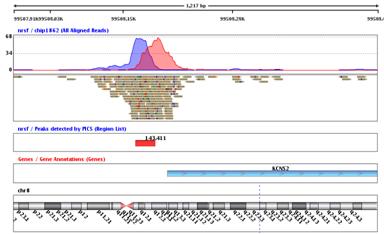
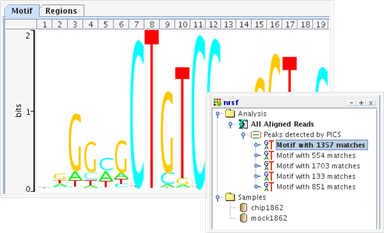
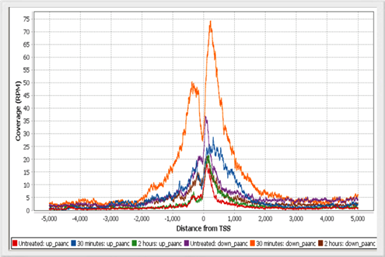
TSS plotVisualize the profile of normalized reads around Transcription Start Sites. |
