|
MeDIP-Seq |
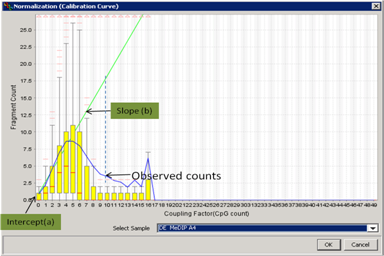
Normalization (Calibration curve)Estimate the linear dependency of methylation signals and CpG counts by calculating the slope and the intercept in a calibration plot. |
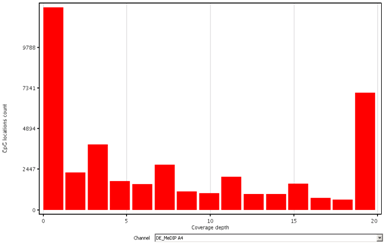
CpG coverage analysisAssess the depth of read coverage across CpG sites. |
|
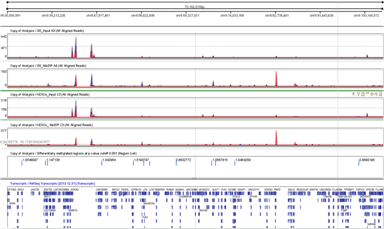
Strand NGS supports genome-wide methylation analysis using MeDIP-Seq data. The workflow supports data normalization before estimating the methylation signal and identifies differential methylation events across a pair of conditions. These regions can be further annotated based on their location with respect to known genes. Downstream analysis such as GO, pathway analysis can be performed on selected entities. |
Differential methylation analysis (DMR)Identify hypomethylated and hypermetylated regions using a p-value threshold. Estimate global and local background noise using input (untreated) samples to help discard false positives. |
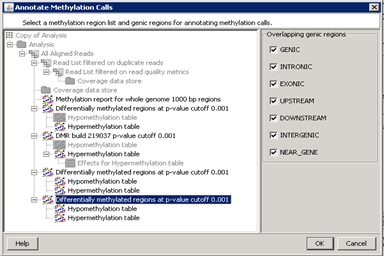
Annotate methylation callsAnnotate DMRs for selected overlapping genic regions based on the chosen genes and transcript model. |